Benchmarking
PyForestScan is designed for high performance and memory efficiency, ensuring it can handle large-scale point cloud datasets effectively. While no other Python libraries specifically calculate these forest structure metrics, there are alternatives in R, such as theleafR library (Almeida et al. 2021), that offer similar functionality.
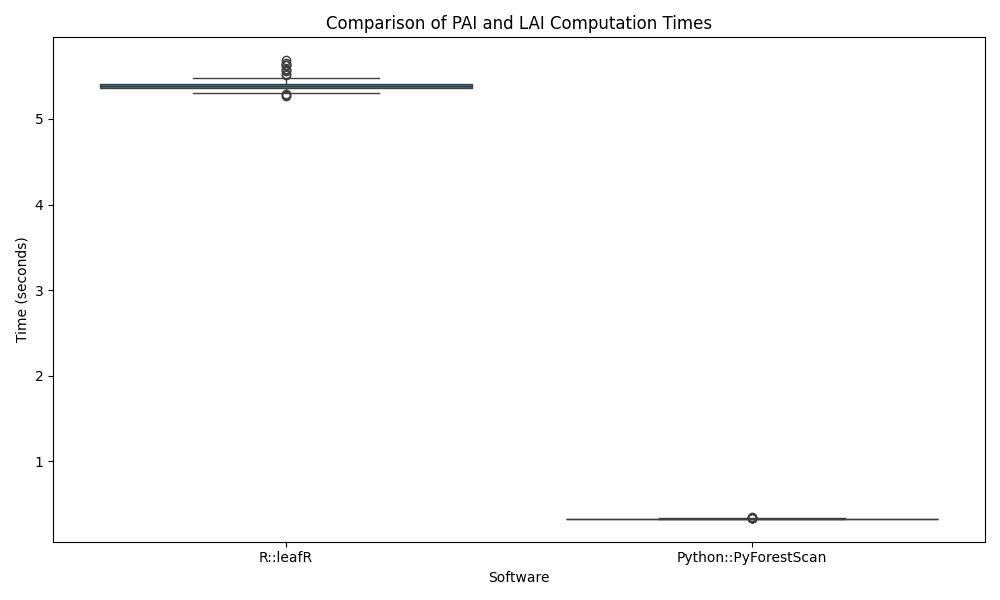
We provide a direct performance comparison between PyForestScan andleafR to demonstrate its efficiency. In both cases, we calculate Plant Area Index (PAI) (this is labelled as Leaf Area Index in the leafR library) on a LAS tile, repeating the process 100 times and plotting the results. The measured benchmarking is only done on the functions to calculate PAI/LAI. It does not include time taken to load the point cloud, etc.
The benchmarks were conducted on a Mac with an Apple M3 Max processor (16 cores) and 128GB RAM.
Code Used
To calculate LAI in leafR, and compare these with the python code, we:
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51 | # Install and load required packages
if (!require("lidR")) install.packages("lidR")
if (!require("raster")) install.packages("raster")
if (!require("leafR")) install.packages("leafR")
library(lidR)
library(raster)
library(leafR)
file_path <- "example_data/20191126_5QKB020840_normalized.laz"
las <- readLAS(file_path)
if (is.empty(las)) {
stop("The LAS file is empty or could not be read.")
}
if (is.null(las@data$Z)) {
stop("The LAS file does not contain Z coordinates.")
}
if (!"Zref" %in% names(las@data)) {
las <- normalize_height(las, tin())
}
temp_las_file <- tempfile(fileext = ".las")
writeLAS(las, temp_las_file)
compute_lai <- function(las_file_path) {
lad_voxels <- lad.voxels(las_file_path, grain.size = 25)
lai_raster <- lai.raster(lad_voxels)
return(lai_raster)
}
timing_results <- data.frame(software = character(),
time = numeric(),
stringsAsFactors = FALSE)
for (i in 1:100) {
start_time <- Sys.time()
lai_result <- compute_lai(temp_las_file)
end_time <- Sys.time()
iteration_time <- as.numeric(difftime(end_time, start_time, units = "secs"))
timing_results <- rbind(timing_results, data.frame(software = "R::leafR", time = iteration_time))
cat(sprintf("Iteration %d completed in %.2f seconds.\n", i, iteration_time))
}
write.csv(timing_results, file = "timing_results.csv", row.names = FALSE)
cat(sprintf("Total time for 100 iterations: %.2f seconds.\n", sum(timing_results$time)))
unlink(temp_las_file)
|
Reference
Almeida, Danilo Roberti Alves de, Scott Christopher Stark, Carlos Alberto Silva, Caio Hamamura, and Ruben Valbuena. 2021. "leafR: Calculates the Leaf Area Index (LAD) and Other Related Functions."Manual. https://CRAN.R-project.org/package=leafR.
